Alias (According to NCBI)
- C-BAS/HAS,
- C-H-RAS,
- C-HA-RAS1,
- CTLO,
- H-RASIDX,
- HAMSV,
- HRAS1,
- K-RAS,
- N-RAS,
- RASH1
- GTP- and GDP-binding peptide B;
- GTPase HRas;
- Ha-Ras1 proto-oncoprotein;
- Ras family small GTP binding protein H-Ras;
- c-has/bas p21 protein;
- c-ras-Ki-2 activated oncogene;
- p19 H-RasIDX protein;
- transformation gene: oncogene HAMSV;
- transforming protein p21
Description
(According to SwissProt)
- Ras proteins bind GDP/GTP and possess intrinsic GTPase activity.
- Chromosomal location
(According to NCBI, CGH: progenetix)
- Location: 11p15.5
- Size: 3309 bp
- exons: 4
- DNA sequence (Human): NC_000011.8
![]()
CGH (11p15.5): Losses (%) -11.7 Gain (%) 2.2
mRNA sequence (Human): Transcript variant1: NM_005343.2
Transcript vairant2:NM_176795.2
Size: Transcript variant1: 1016 bp
Transcript variant2: 1143 bp
cDNA libraries:
- Size: 189 amino acids: 21298 Da
Subcellular location: Cell membrane; Lipid-anchor; Cytoplasmic side. Golgi apparatus membrane; Lipid-anchor. Note=Shuttles between the plasma membrane and the Golgi apparatus.
Protein domains

Protein sequence (Human): P01112
Homologous genes: 55890
2D PAGE:
3D Structure: 121P 1AA9 1AGP 1BKD 1CLU 1CRP 1CRQ 1CRR 1CTQ 1GNP 1GNQ 1GNR 1HE8 1IAQ 1IOZ 1JAH 1JAI 1K8R 1LF0 1LF5 1LFD 1NVU 1NVV 1NVW 1NVX 1P2S 1P2T 1P2U 1P2V 1PLJ 1PLK 1PLL 1Q21 1QRA 1RVD 1WQ1 1XCM 1XD2 1XJ0 1ZVQ 1ZW6 221P 2C5L 2CE2 2CL0 2CL6 2CL7 2CLC 2CLD 2EVW 2GDP 2Q21 2QUZ 2RGA 2RGB 2RGC 2RGD 2RGE 2RGG 2UZI 2VH5 421P 4Q21 521P 5P21 621P 6Q21 721P 821P
PTM:
Palmitoylated by the ZDHHC9-GOLGA7 complex. A continuous cycle of de- and re-palmitoylation regulates rapid exchange between plasma membrane and Golgi.
S-nitrosylated; critical for redox regulation. Important for stimulating guanine nucleotide exchange. No structural perturbation on nitrosylation.
Pathway:
hsa04010 MAPK signaling pathway
hsa04012 ErbB signaling pathway
hsa04360 Axon guidance
hsa04370 VEGF signaling pathway
hsa04510 Focal adhesion
hsa04530 Tight junction
hsa04540 Gap junction
hsa04650 Natural killer cell mediated cytotoxicity
hsa04660 T cell receptor signaling pathway
hsa04662 B cell receptor signaling pathway
hsa04664 Fc epsilon RI signaling pathway
hsa04720 Long-term potentiation
hsa04730 Long-term depression
hsa04810 Regulation of actin cytoskeleton
hsa04910 Insulin signaling pathway
hsa04912 GnRH signaling pathway
hsa04916 Melanogenesis
hsa05211 Renal cell carcinoma
hsa05213 Endometrial cancer
hsa05214 Glioma
hsa05215 Prostate cancer
hsa05216 Thyroid cancer
hsa05218 Melanoma
hsa05219 Bladder cancer
hsa05220 Chronic myeloid leukemia
hsa05221 Acute myeloid leukemia
hsa05223 Non-small cell lung cancer
Protein interactions: HRAS
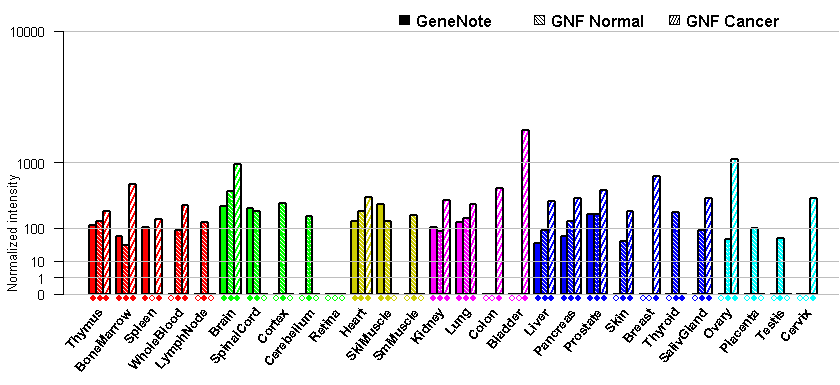
- Tissue expression: PubMed Reference
- Human Protein Atlas (HPA): CAB002015
- OMIM : 190020
- PubMed: Early detection Diagnosis Prognosis Therapy