Alias (According to NCBI)
- cytochrome P450, family 3, subfamily A, polypeptide 5 pseudogene 2
- cytochrome P450, subfamily IIIA, polypeptide 5 pseudogene 2
- cytochrome P450, subfamily IIIA, pseudogene 2
Description
(According to SwissProt)
Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics.
- Chromosomal location
(According to NCBI, CGH: progenetix)
- Location: 7q21.3-q22.1
- Orientation: minus strand
- Size: 4,242 bp
- 2 exons
- DNA sequence (Human): NC_000007
![]()
CGH (7q21.3 - 22.1): Losses (%) - 2.8 Gain (%) 11.2
- HGMD:
- SNPs: CYP3A5P2
mRNA sequence (Human):
Size:
cDNA libraries:
- Size: 502 a.a., 57,109 Da
- Catalytic activity:RH + reduced flavoprotein + O(2) = ROH + oxidized flavoprotein + H(2)O.
Subcellular location: RH + reduced flavoprotein + O(2) = ROH + oxidized flavoprotein + H(2)O.
Protein domains
Protein sequence (Human): P20815
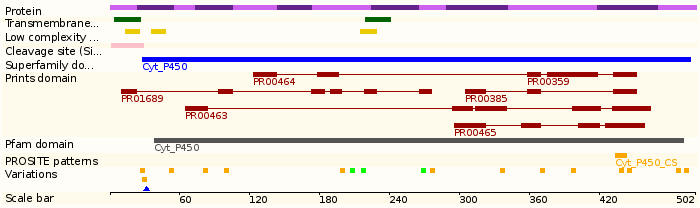
Homologous genes:
2D PAGE:
3D Structure:
PTM:
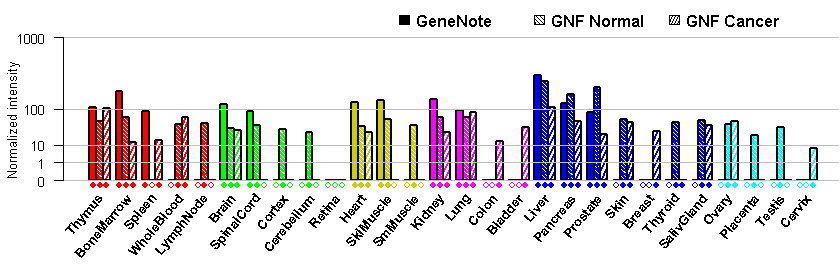
- Tissue expression: PubMed Reference
- Human Protein Atlas (HPA):
OMIM:
PubMed: Early detection Diagnosis Prognosis Therapy