 Version
II
Version
II Home
| Gene list | Search
Home
| Gene list | Search
 Version
II
Version
II Home
| Gene list | Search
Home
| Gene list | Search
Total 374 genes in the database are categorized into
biological processes and molecular functions based on
Gene Ontology using
FATIGO [
Al-Shahrour F., D�az-Uriarte R., &
Dopazo J.,
FatiGO: a web tool for finding significant associations of Gene Ontology terms
with groups of genes, Bioinformatics, 20, 578-580, 2004
]
Genes belonging to major biological processes and molecular
functions are listed in table below.
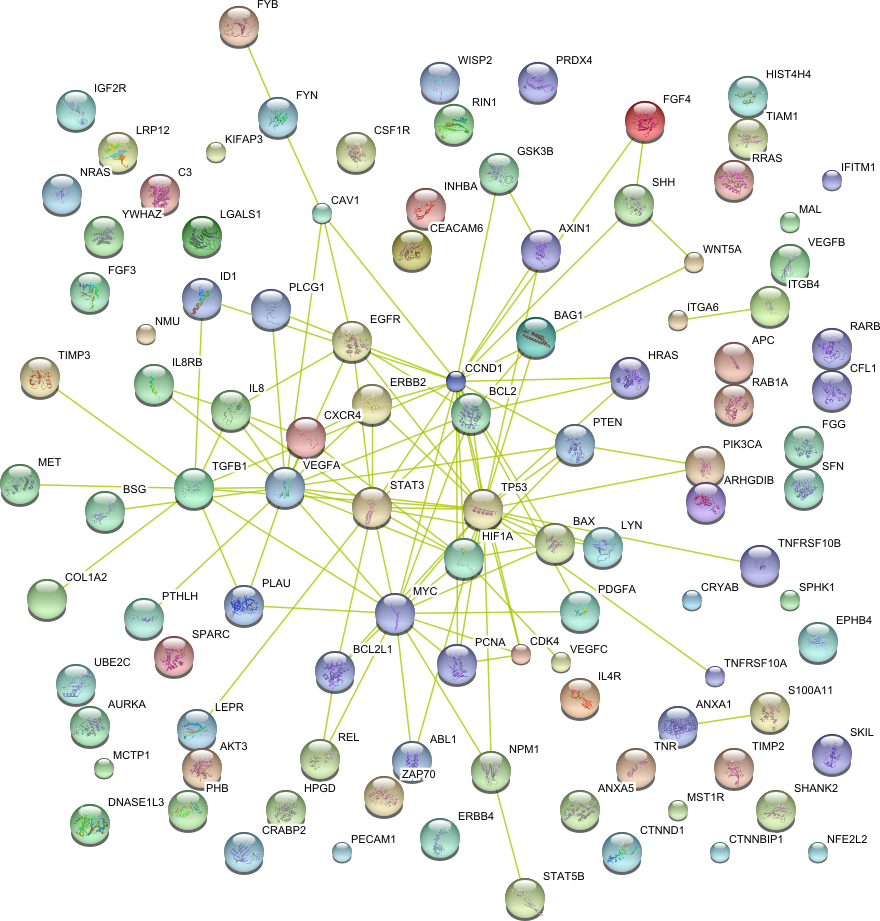
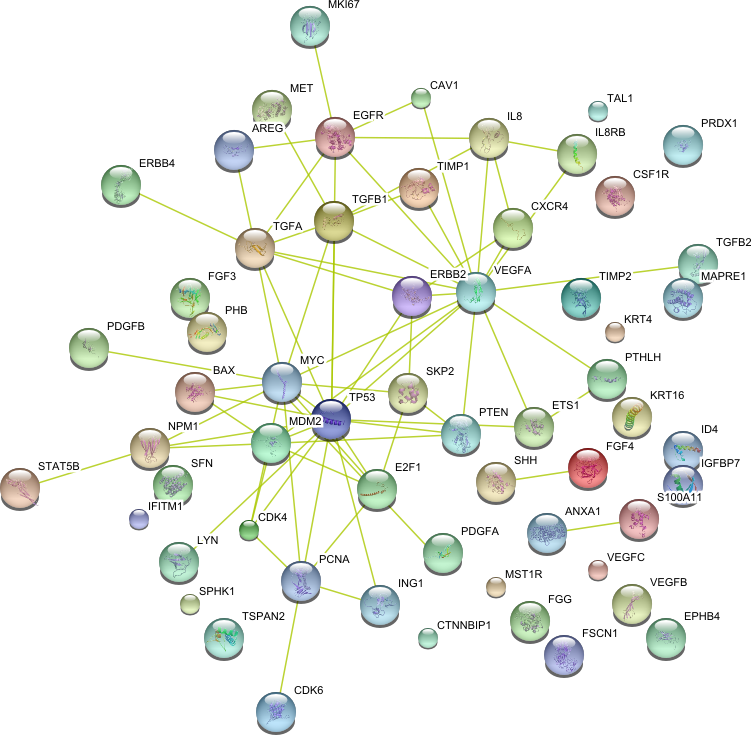
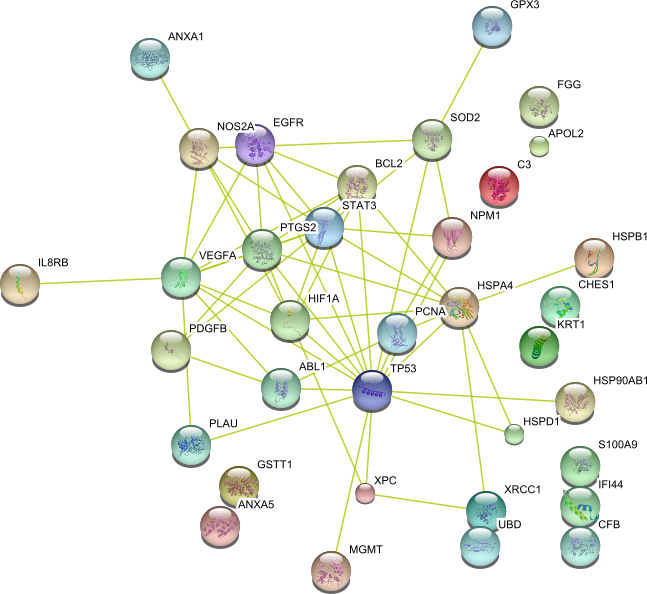
String 8.3 tool has been used to predict protein-protein interaction networks. [
STRING 8--a global view on proteins and their functional interactions in 630
organisms
Nucleic
Acid Research, 37, D412 - 6, 2009 ]
Gene Ontology :- Biological Process
| Biological Process | Genes | No. of genes |
Interaction Network (Download SVG Viewer to view the link enable Map ) |
| Signal transduction (GO:0007165) | CCND1 FYN ANXA5 ARHGDIB SPARC PDGFA TNFRSF10A CAV1 FGF4 CEACAM6 MET ITGA6 TIAM1 AXIN1 COL1A2 ABL1 WISP2 CXCR4 CRABP2 IL8RB SHANK2 YWHAZ SHH LGALS1 VEGFC CRYAB HPGD PTEN PTHLH PRDX4 TIMP3 BSG NMU WNT5A HIST4H4 MCTP1 HIF1A FYB KIFAP3 NPM1 ITGB4 EPHB4 CDK4 APC ANXA1 SFN VEGFA S100A11 IL4R UBE2C TNFRSF10B CFL1 PLCG1 MYC REL ZAP70 LRP12 PCNA DNASE1L3 FGG CTNNBIP1 IL8 BAG1 TGFB1 Hras LEPR MAL IFITM1 BCL2 NFE2L2 AKT3 RRAS IGF2R ErbB2 STAT5B CTNND1 PHB C3 TP53 LYN NRAS INHBA RAB1A GSK3B HRAS ERBB4 FGF3 RIN1 EGFR VEGFB PLAU Bax TNR STAT3 SPHK1 PECAM1 BCL2L1 TIMP2 ID1 MST1R PIK3CA SKIL CSF1R RARB AURKA | 105 |
 |
| Cell proliferation (GO:0008283) |
PTEN
PTHLH
FSCN1
NPM1
EPHB4
CDK4
ANXA1
SFN
VEGFA
S100A11
PDGFB
KRT4
MYC
PCNA
FGG
ETS1
CTNNBIP1
IL8
TGFB1
AREG
TIMP1
SKP2
IFITM1
TSPAN2
ErbB2
STAT5B
PHB
ING1
TP53
MDM2
LYN
ERBB4
FGF3
EGFR
VEGFB
Bax
E2F1
SPHK1
TGFB2
KRT16
TIMP2
MKI67
MST1R
CSF1R
TGFA
TAl1
PDGFA
CAV1
FGF4
IGFBP7
MET
CDK6
ID4
PRDX1
CXCR4
IL8RB
MAPRE1
SHH
VEGFC
|
59 |
 |
| Response to stress (GO:0006950) | CLU ABL1 GSTT1 CXCR4 IL8RB MGMT SPRR3 GPX3 HIF1A KRT1 NPM1 S100A9 ANXA1 SFN VEGFA XRCC1 PDGFB PCNA FGG IL8 TGFB1 PRDX2 CHES1 HSPB1 ALB XPC NFE2L2 STAT5B MSH2 S100A7 C3 TP53 SERPINE1 SOD2 HSPD1 EGFR PLAU Bax OGG1 STAT3 PECAM1 TGFB2 MICA CCND1 ANXA5 APOL2 FOS HSPA1A | 48 |
 |
| AXIN1 CLU ABL1 CXCR4 YWHAZ BIRC5 LGALS1 CTSB MGMT PTEN HIF1A NPM1 ANXA1 SFN VEGFA TNFRSF10B CFL1 MYC PEG3 DNASE1L3 BAG1 PRDX2 MMP9 HSPB1 MAL ALB BCL2 STAT5B MSH2 PHB TP53 INHBA HSPD1 GSTP1 Bax TUBB2C E2F1 SPHK1 BCL2L1 TGFB2 PIK3CA ANXA5 TNFRSF10A HSPA1A FGF4 | 45 |
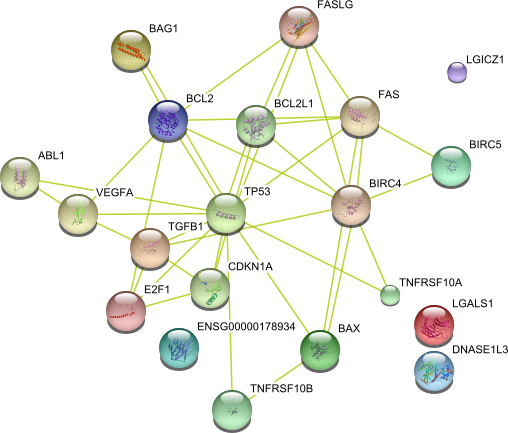 |
|
| Cell adhesion (GO:0007155) | DSG3 ARHGDIB CD44 FAT ICAM1 ITGA6 SNAI1 VCAN ABL1 WISP2 DSG1 APBA1 ATP2A2 CDH2 LAMC2 LAMA5 PTEN ITGB4 APC POSTN IL8 TSPAN2 STAT5B CTNND1 CDH3 CLDN17 EGFR CDH1 CDH17 TNR PECAM1 PKP3 | 32 |
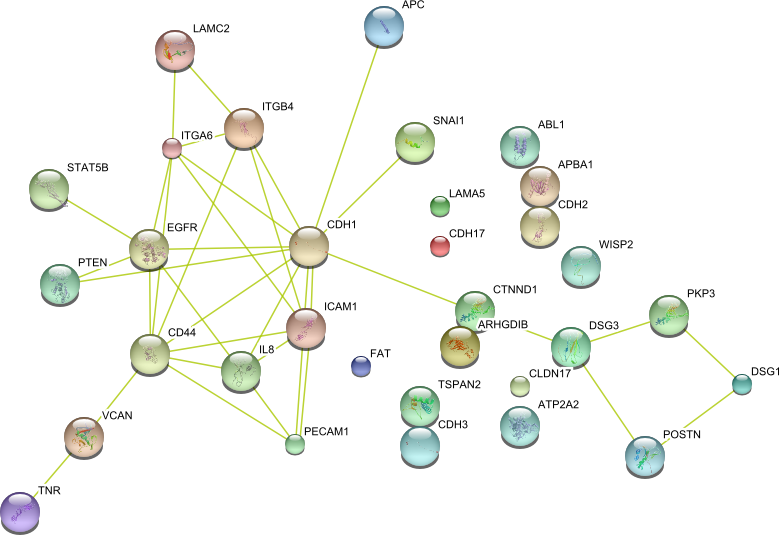 |
| Angiogenesis (GO:0001525) | SPHK1 TGFB2 ID1 TGFA PDGFA CXCR4 SHH VEGFC HIF1A KRT1 EPHB4 VEGFA IL8 ErbB2 S100A7 SERPINE1 SPINK5 | 17 |
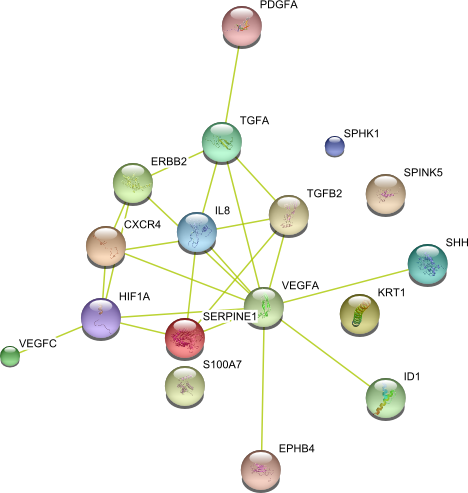 |
|
Molecular Function |
Genes |
No. of genes |
Interaction Network (Download SVG Viewer to view the link enable Map ) |
| Ion binding | HEXA MMP9 ITGA6 DSG3 S100A7 S100A11 TCF20 S100A9 ZNF273 STAT3 CYP2D6 FGG VCAN CYP1A1 CYP2E1 SCEL DNASE1L3 COPS5 TP53 ALB ENO1 MSH2 STAT5B NPR3 PKM2 ATP2A2 PLCG1 CDH17 MYL1 SOD2 ATP5B HBA1 FXYD3 S100P PPP1CA FHIT MCM2 MMP1 ABL1 BIRC5 FAT MCTP1 CA2 CDH1 CDH2 CDH3 SPARC SNAI1 RARB WT1 BMI1 PEG3 MMP10 COX1 HBB MMP11 COX2 ANXA1 ANXA2 MGMT MMP2 MDM2 TGM3 MMP3 MMP14 PTHLH ANXA5 S100A2 DZIP1 SPHK1 S100A4 MMP7 DSG1 ING1 | 74 |
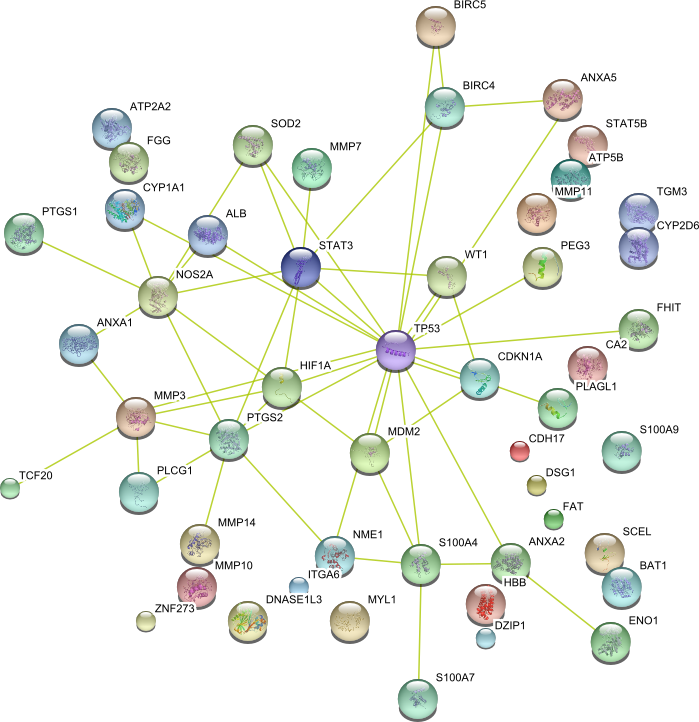 |
| Nucleic acid binding (GO:0003676) | SIP1 SMARCA4 FOS EIF4E MCM2 HIF1A NPM1 ABL1 USF1 PCNA NRAS HIST4H4 SNAI1 RARB WT1 PEG3 MGMT HOXA1 OGG1 H3F3A BAT1 DZIP1 SPHK1 CHES1 ETV4 SKIL NFE2L2 XPC TCF20 HOP ZNF273 STAT3 EEF1A1 REL DNASE1L3 COPS5 MYC TP53 ALB ENO1 MSH2 STAT5B JUN TERT EGFR E2F1 ETS1 TAl1 HMGA2 XRCC1 | 50 |
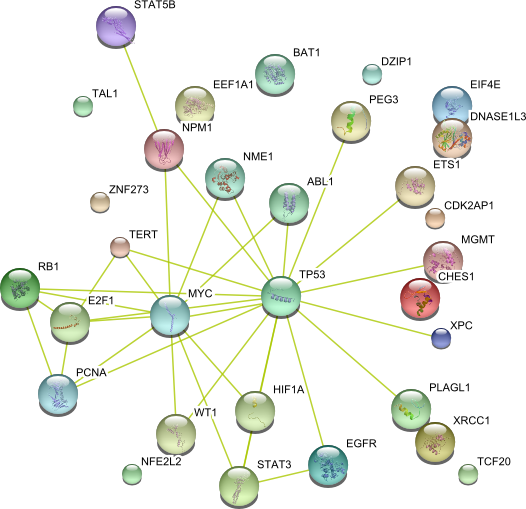 |
| hydrolase activity (GO:0016787) | PPP1CA FHIT SMARCA4 PTEN HGF MCM2 RRAS CHKA MMP1 NRAS TUBB2C MMP10 MMP2 OGG1 ABCB1 TGM3 SHH MMP3 MMP14 P11 BAT1 ABCB4 MMP7 PPP1R3C CTSB HEXA MMP9 RAB1A PLAU EEF1A1 SPINK5 SERPINE1 DNASE1L3 TP53 ENO1 MSH2 CTSD ATP2A2 PGAM1 PTPN13 PLCG1 CES2 ATP5B EPHX1 CYLD | 45 |
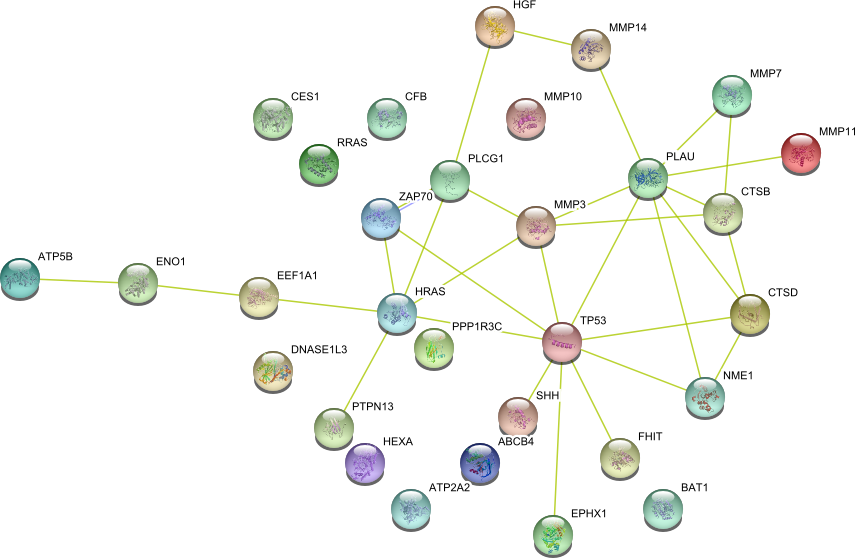 |
| Transferase activity (GO:0016740) | GSTP1 EPHB4 TK1 CHKA GSK3B ABL1 PCNA ERBB4 CSF1R MST1R MGMT FYN TGM3 ZAP70 SPHK1 CKM CCND1 CDK4 PLAU MET GSTT1 PAPSS2 ErbB2 GSTM1 PIK3CA GSTM3 MSH2 AKT3 CDK6 PKM2 TERT EGFR LYN AURKA AURKB | 35 |
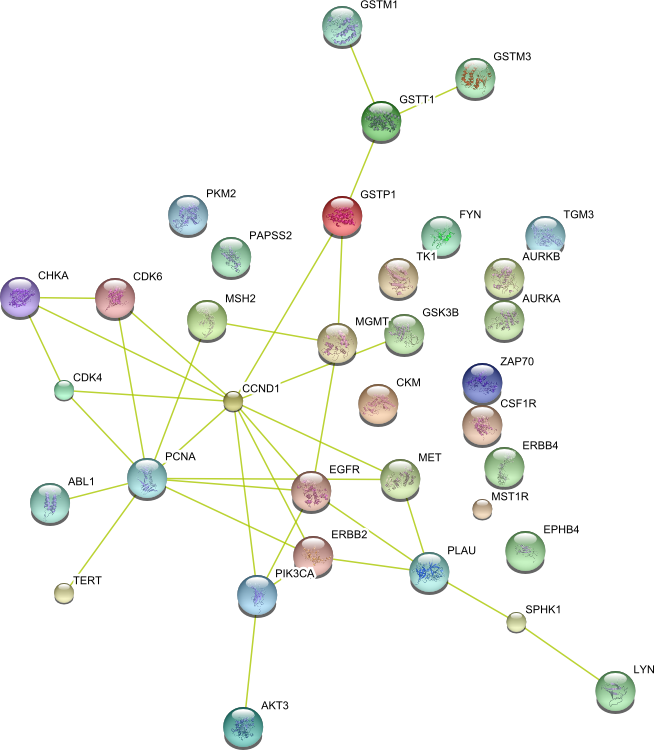 |
| Receptor binding (GO:0005102) | IK PDGFA FYB LYN PDGFB FGF3 TGFA LAMA5 HGF C3 NMU FGF4 TUBB2C APOL2 WNT5A ANXA1 SHH DBI MSN PTHLH IL8 AREG P11 TIMP2 INHBA VEGFA FGG VEGFB VEGFC ErbB2 TNR TGFB1 NRG1 TGFB2 TIAM1 | 35 |
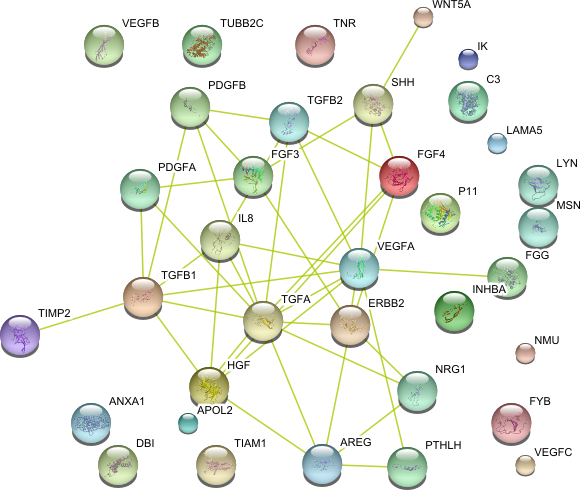 |