DOME execution
For running any script pertaining to DOME, make sure that the domeenv Conda environment is activated. More information about Conda environments can be found here
Running DOME command line (CLI) mode
The required scripts to run DOME are placed in the “script” directory.
To run DOME as a command line program, use the following commane: In Linux terminal:
$ conda activate domeenv
$ cd DOME-main/scripts
$ python3 metaaccesor.py <input_csv>
Format of the above mentioned "input_csv" should be as shown below. The CSV format file should contain 3 columns (column 1 - uniprot entry name of the protein; column 2 - amino acid position; column 3 - ALTERED amino acid) and may contain multiple rows (user defined). An example of the file contents is shown below :
EGFR_HUMAN,858,R
ERBB4_HUMAN,931,Y
EGFR_HUMAN,829
TF7L2_HUMAN,365
EGFR_HUMAN,790,MRunning DOME-GUI
To load the R Shiny based GUI or via web-browser use the following command:
In Linux terminal:
$ conda activate domeenv
$ R -e "shiny::runApp('app.R', launch.browser = TRUE)"
In Windows CMD:
> "Path/to/R.exe" -e "shiny::runApp("app.R", launch.browser = TRUE)"
Using DOME-GUI
Once users executes the above commands DOME GUI appears. DOME-GUI can be used for multiple queries (as in case of CLI mode) or explore individual protein positions.
Following are the detailed description for each of the tabs-
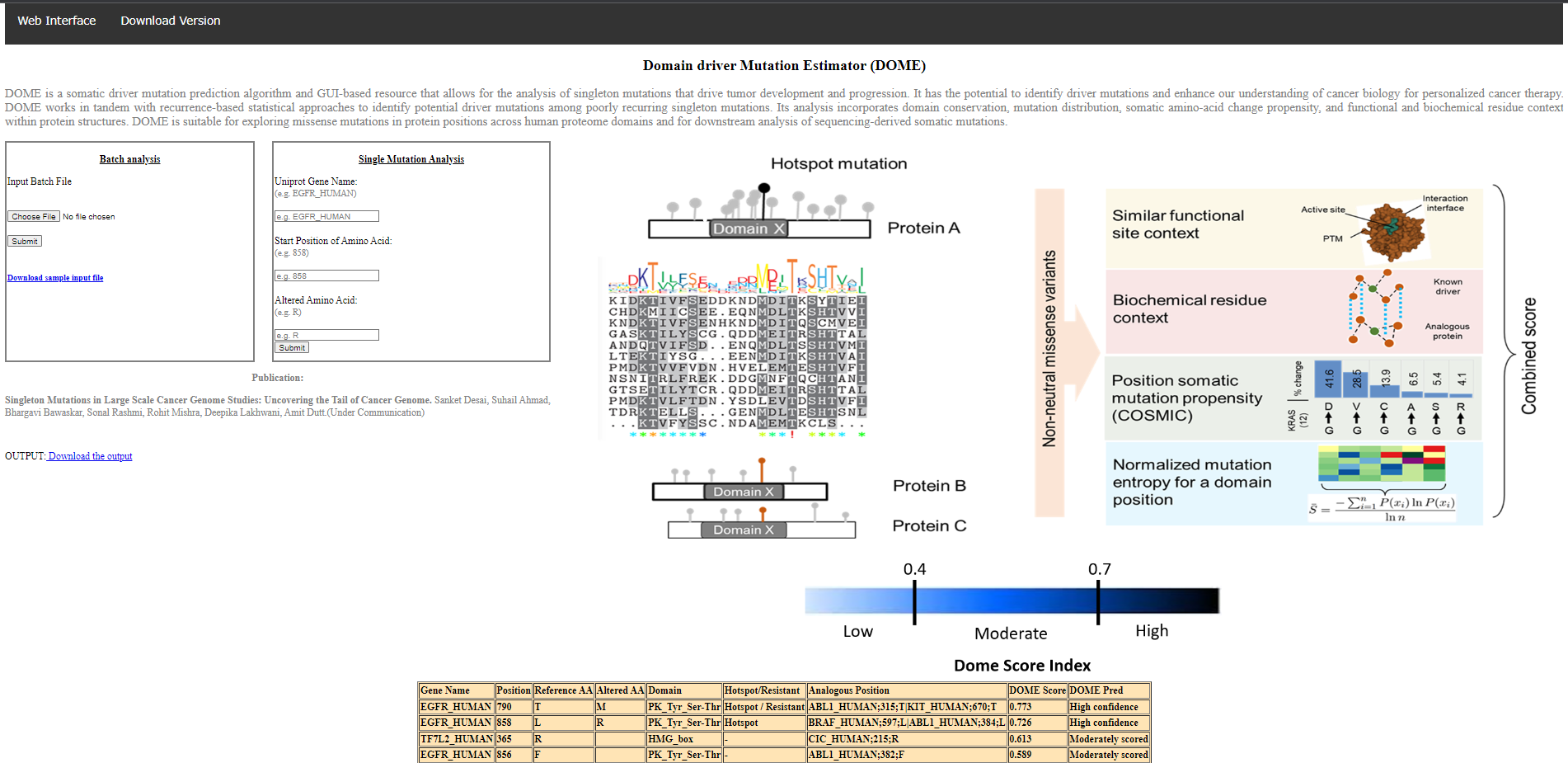
For multiple queries - user may load the input file (in the CSV format shown above) and upon clicking "Submit", the results would be presented in a tabular Format as shown in below:
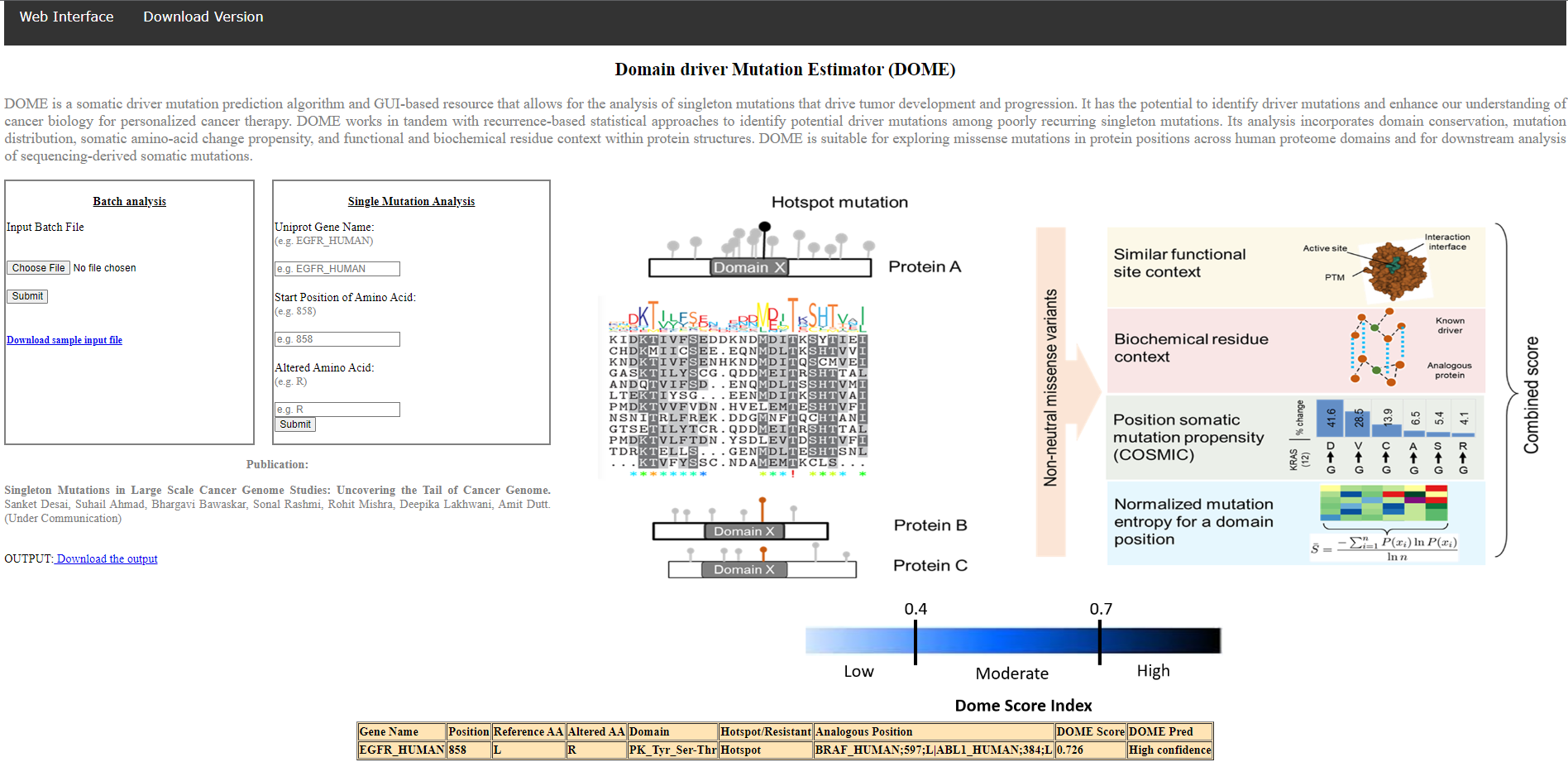
For individual protein queries - user may input the protein name (Uniprot entry name), position (range with start and end position) and the results for the prediction would be presented in a tabular Format as shown in below:
DOME output
Gene based or mutation (position) based search returns a table containing the following columns:
| Column names | Description |
|---|---|
| Entry name | Entry name of protein as per Uniprot database |
| Position | Amino acid position in the protein |
| Reference | Reference amino acid in the protein |
| Altered | Altered amino acid in the protein |
| Domain | Domain name |
| Mutation type | H - hotspot, R - resistant, '-' - not present in significant mutation database |
| Analogous to | Query protein position is analogous to this known significant mutation |
| DOME Score | Score computed using the DOME algorithm scoring metric (scaled 0-1) |