Mass Spectrometry
Overview
The Mass Spectrometry facility at ACTREC houses four state-of-the-art mass spectrometry platforms connected to high performance liquid chromatography systems – (1) Orbitrap Exploris 480, Thermo, (2) ESI/MALDI Q-TOF Model Synapt XS (3) Triple Quadrupole Model Xevo TQ Absolute and (4) The Nano-LC (ABSCIEX, Eksigent)-ESI-Q-TOF Model Triple TOF 5600 plus SCIEX.
Mass spectrometry is a technique which measures mass-to-charge ratios of ions. This can be used to identify and quantify the molecules of interest in the given sample. Since the advent of soft ionization methods, large biomolecules like proteins are increasingly being analyzed by mass spectrometry for studying various aspects of their structure. Mass spectrometry based Proteomics, i.e., large scale identification and quantification of proteins from a complex mixture of biological origin; and variations thereof, has become an important workflow in biological research. High resolution mass spectrometry is primarily applied for large scale discovery studies while low resolution mass spectrometry is more suited for targeted proteomics. At ACTREC, we have three high resolution mass spectrometers (MS) and one low resolution MS dedicated for omics applications.
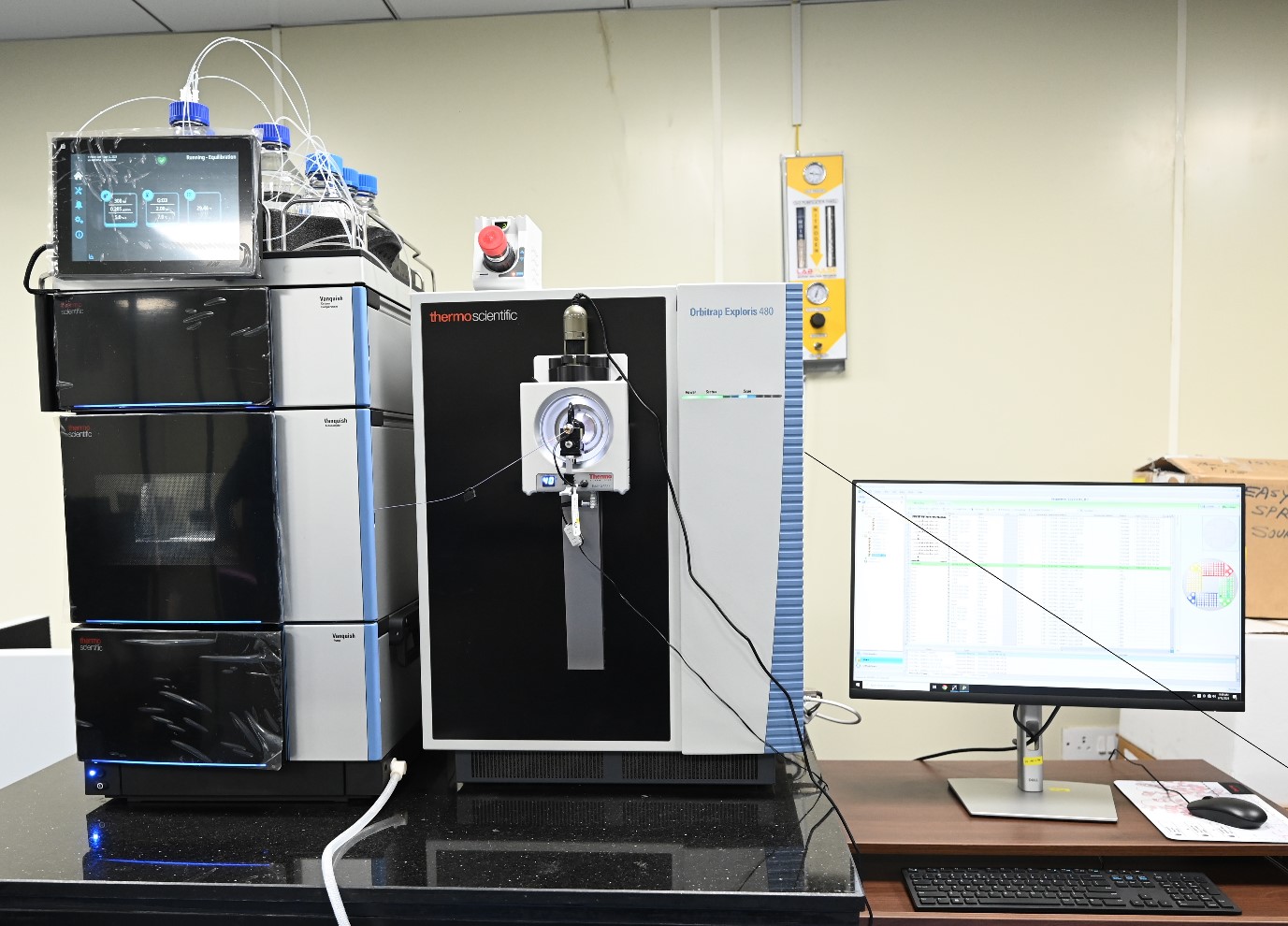

Orbitrap Exploris 480, Thermo
This high resolution MS system has an ESI ion source and a nanoFlex ion source and the mass analyzer is an Orbitrap. This system is coupled with a Vanquish nanoLC. The high resolution and high sensitivity provided by this system is useful in shotgun proteomics applications of discovery nature. There are specialized data analysis software suits for analysis of the raw data, namely Proteome Discoverer and Spectranaut. This system is well known for in-depth proteomic profiling of the sample, label and label-free quantification of the proteins in a complex mixture. This is well suited for comparative proteomics studies leading to biomarker identification for various disease conditions etc.

Synapt XS, Waters
This is a high resolution MS with an interesting option of changing the ion sources from ESI, nanoESI and MALDI (matrix assisted laser desorption ionization). For ESI and nanoESI application, it can be coupled with the M-Class UPLC from Waters and can be used for various shotgun proteomics, lipidomics, metabolomics applications. The main mass analyzer is TOF and the system is equipped with Ion Mobility Separation of the ions, providing separation between molecules of same molecular weight but different conformations. Since this system also has an option of Electron Transfer Dissociation (ETD), this is suitable for identification and characterization of the post-translational modifications (PTMs). With MALDI as an ion source, this system can be utilized for tissue MALDI imaging. The data analysis software available is Progenesis QIP and MassLynx.

Xevo TQ Absolute, Waters
This is a low resolution MS system with three quadrupole mass analyzers in series. It is coupled with an H-Class UPLC and has an ESI ion source. This system is best suited for targeted proteomics studies, where absolute or relative quantification of the protein/peptide/small molecule of interest can be done using single or multiple reactions monitoring technique (SRM/ MRM) and its variations. The unparalleled sensitivity provided by the MRM assay can be used to track extremely low amounts of compounds of interest in the given sample. The analysis software MassLynx software helps in analyzing the data of targeted quantification experiments

TripleTOF 5600 plus, SCIEX
This is a high resolution MS with nanoESI (electrospray ionization) source and nanoLC coupled with it. The main mass analyzer is Time-of-Flight (TOF). This system is useful for shotgun proteomic experiments of qualitative and quantitative nature. The patented SWATH technology of Sciex helps in increasing the depth of the proteomic identification as well as label-free quantification, and thereby able to identify the proteins with very low expression. Along with that, the system can also be used for label-based quantification of proteins. The analysis software ProteinPilot is useful for qualitative and quantitative proteomics studies.| Sr. No. | Applications |
|---|---|
| 1 | Enriched Proteins (IP / In-gel digested samples) |
| 2 | Global Protein Profiling (Complex Mixture) |
| 3 | Label Free Quantification |
| 4 | Labelled Quantification (i-TRAQ / TMT) |
| 5 | Parallel Reaction Monitoring (PRM) |
| 6 | PTMs |
| 7 | Metabolomics |
| 8 | Lipidomics |
| 9 | MALDI MS |
| 10 | MALDI MS-MS |
| 11 | Targeted Proteomics Studies |
| 12 | SRM / MRM studies |
2025
- ATP-Driven Allosteric Regulation of 14-3-3:PositiveModulation of ATP Hydrolysis and Negative Regulation of Peptide Binding
- Neha, A., and G. Rukmini. 2025. “Incremental Modification in the Existing Approaches for Affinity Chromatographic Enrichment of Phosphoproteins Improves Their Profile in Liquid Chromatography‐Tandem Mass Spectrometry Analysis.” Analytical Science Advances 6(1).
- Chaudhary, N., B. S. Choudhary, A. Shivashankar, S. Manna, K. Ved, S. Shaikh, and N. Verma. 2025. “EGFR-to-Src Family Tyrosine Kinase Switching in Proliferating-DTP TNBC Cells Creates a Hyperphosphorylation-Dependent Vulnerability to EGFR TKI.” Cancer Cell International 25 (1):55.
2023
2022
- Nimbalkar, Vaishnavi K., Jeet Gangar, Saptarsi Shai, Pallavi Rane, Subham Kumar Mohanta, Sadhana Kannan, Arvind Ingle, Neha Mittal, Swapnil Rane, and Manoj B. Mahimkar. 2022. “Prevention of Carcinogen-Induced Oral Cancers by Polymeric Black Tea Polyphenols via Modulation of EGFR-Akt-MTOR Pathway.” Scientific Reports 12(1). doi: 10.1038/s41598-022-18680-0.
- Narasimhan, Mythreyi, Vaishnavi Khamkar, Sarika Tilwani, Sorab N. Dalal, Dhanlaxmi Shetty, P. G. Subramanian, Sanjay Gupta, and Rukmini Govekar. 2022. “Atypical Activation of Signaling Downstream of Inactivated Bcr-Abl Mediates Chemoresistance in Chronic Myeloid Leukemia.” Journal of Cell Communication and Signaling 16(2). doi: 10.1007/s12079-021-00647-x.
- Sreevidya, T. S., Somavally Dalvi, Prasanna Venkatraman, and Satyavani Vemparala. 2022. “Structural Insights on the Effects of Mutation of a Charged Binding Pocket Residue on Phosphopeptide Binding to 14-3-3ζ Protein.” Proteins: Structure, Function and Bioinformatics 90(5).
- Deo, Abhilash Nitin, Rahul Thorat, Ajit Chandrakant Dhadve, Abhijit De, Bharat Rekhi, and Pritha Ray. 2022. “IGF1R-Α6 Integrin-S100A4 Network Governs the Organ-Specific Metastasis of Chemoresistant Epithelial Ovarian Cancer Cells.” Biochimica et Biophysica Acta - Molecular Basis of Disease 1868(1).
- Govekar, Rukmini. 2022. “Improved Identification of Hydrophobic Proteins by Optimization of LC Conditions within the LC-MS Run: A Practical Strategy for Scanty Clinical Samples".” Biomedical Journal of Scientific & Technical Research 43(4).
2021
- Rajendra, Jacinth, Atanu Ghorai, and Shilpee Dutt. 2021. “14-3-3ζ Negatively Regulates Mitochondrial Biogenesis in GBM Residual Cells.” Heliyon 7(11).
- Ud Din Farooqee, Sheikh Burhan, Joel Christie, and Prasanna Venkatraman. 2021. “PSMD9 Ribosomal Protein Network Maintains Nucleolar Architecture and WT P53 Levels.” Biochemical and Biophysical Research Communications 563
- Das, L., V. Murthy, and A. K. & VARMA. 2021. “Quantitative Serum Proteomics Reveals The Predictive and Prognostic Potential of Clusterin and Gelsolin in Head and Neck Squamous Cell Carcinoma Treated With Radiotherapy.” (Pre-Print on Research Square).
2020
2019
- Narasimhan, Mythreyi, Sadhana Kannan, Aakash Chawade, Atanu Bhattacharjee, and Rukmini Govekar. 2019. “Clinical Biomarker Discovery by SWATH-MS Based Label-Free Quantitative Proteomics: Impact of Criteria for Identification of Differentiators and Data Normalization Method.” Journal of Translational Medicine 17(1).
- Sankaran, H., S. Sengupta, V. Purohit, A. Kotagere, N. R. Moulik, M. Prasad, and V. Gota. 2019. “A Comparison of Asparaginase Activity in Generic Formulations of E. Coli Derived L‐asparaginase: In‐vitro Study and Retrospective Analysis of Asparaginase Monitoring in Pediatric Patients with Leukemia.” British Journal of Clinical Pharmacology 86(6):1081–88.
2018
- Jain, Bhawik Kumar, Pankaj Singh Thapa, Ashok Varma, and Dibyendu Bhattacharyya. 2018. “Identification and Characterization of GRIP Domain Golgin PpImh1 from Pichia Pastoris.” Yeast 35(8).
- Nishad, Srambikkal, and Anu Ghosh. 2018. “Comparative Proteomic Analysis of Human Peripheral Blood Mononuclear Cells Indicates Adaptive Response to Low-Dose Radiation in Individuals from High Background Radiation Areas of Kerala.” Mutagenesis 33(5–6).
2017
- Hudlikar, Rasika R., Varadha Balaji Venkadakrishnan, Rajiv Kumar, Rahul A. Thorat, Sadhana Kannan, Arvind D. Ingle, Saral Desai, Girish B. Maru, and Manoj B. Mahimkar. 2017. “Polymeric Black Tea Polyphenols (PBPs) Inhibit Benzo(a)Pyrene and 4-(Methylnitrosamino)-1-(3-Pyridyl)-1- Butanone-Induced Lung Carcinogenesis Potentially through down-Regulation of P38 and Akt Phosphorylation in A/J Mice.” Molecular Carcinogenesis 56(2)
- Bhattacharya, Saikat, Divya Reddy, Vinod Jani, Nikhil Gadewal, Sanket Shah, Raja Reddy, Kakoli Bose, Uddhavesh Sonavane, Rajendra Joshi, and Sanjay Gupta. 2017. “Histone Isoform H2A1H Promotes Attainment of Distinct Physiological States by Altering Chromatin Dynamics.” Epigenetics and Chromatin 10(1).
- Yadav, Lumbini R., Mahamaya N. Biswal, M. v. Hosur, Nachimuthu Senthil Kumar, and Ashok K. Varma. 2017. “Structural Basis to Characterise Transactivation Domain of BRCA1.” Journal of Biomolecular Structure and Dynamics 35(1).
- Thombre, Rebecca, Vinaya Shinde, Jyotsana Dixit, Sagar Jagtap, and Pandit B. Vidyasagar. 2017. “Response of Extreme Haloarchaeon Haloarcula Argentinensis RR10 to Simulated Microgravity in Clinorotation.” 3 Biotech 7(1).
2016
- Thombre, Rebecca S., Vinaya D. Shinde, Radhika S. Oke, Sunil Kumar Dhar, and Yogesh S. Shouche. 2016. “Biology and Survival of Extremely Halophilic Archaeon Haloarcula Marismortui RR12 Isolated from Mumbai Salterns, India in Response to Salinity Stress.” Scientific Reports 6
- Chavan, Rahul, Sandeepan Mukherjee, Ritwik Dahake, Domnic Colvin, Avinash Kale, and Abhay Chowdhary. 2016. “Differential Proteomic Analysis of Respiratory Samples from Patients Suffering from Influenza.” VirusDisease 27(3).
- Nishad, S., and A. Ghosh. 2016. “Dynamic Changes in the Proteome of Human Peripheral Blood Mononuclear Cells with Low Dose Ionizing Radiation.” Mutation Research - Genetic Toxicology and Environmental Mutagenesis 797
- Mukhopadhyay, Amitabha, Lalit Sehgal, Arunabha Bose, Anushree Gulvady, Parijat Senapati, Rahul Thorat, Srikanta Basu, Khyati Bhatt, Amol S. Hosing, Renu Balyan, Lalit Borde, Tapas K. Kundu, and Sorab N. Dalal. 2016. “14-3-3γ Prevents Centrosome Amplification and Neoplastic Progression.” Scientific Reports 6
- Bhanushali, Paresh B., Shamkant B. Badgujar, Mukesh M. Tripathi, Sanjeev Gupta, Vedang Murthy, Musti V. Krishnasastry, and Chander P. Puri. 2016. “Development of Glycan Specific Lactin Based Immunoassay for Detection of Prostate Specific Antigen .” International Journal of Biological Macromolecules 86:468–80
2015
- Sonani, Ravi Raghav, Mahima Sharma, Gagan Deep Gupta, Vinay Kumar, and Datta Madamwar. 2015. “Phormidium Phycoerythrin Forms Hexamers in Crystals: A Crystallographic Study.” Acta Crystallographica Section:F Structural Biology Communications 71
- Darshana Salaskar, Sharad P. Kale. 2015. “Isolation and Identification of Arsenic Resistant Providencia Rettgeri (KDM3) from Industrial Effluent Contaminated Soil and Studies on Its Arsenic Resistance Mechanisma.” Journal of Microbial & Biochemical Technology 07(04).
- Rebecca S Thombre, and Radhika S Oke. 2015. “Study of Stress Protein Induced by Temperature Stress in Extremely Halophilic Archaea, Haloferax Mediterranei RT18.” Int J Curr Microbiol App Sci 2:199–209.
2014
- Sharma, Savita, Vinny Punjabi, Surekha M. Zingde, and Sadashiv M. Gokhale. 2014. “A Comparative Protein Profile of Mammalian Erythrocyte Membranes Identified by Mass Spectrometry.” Journal of Membrane Biology 247(11).
- Gore, Manish, and Rukmini Govekar. 2014. “Preliminary Assessment on Choice of Matrix Based on Hydrophobicity/Hydrophilicity of Proteins in Analysis by MALDI Tof-Tof Mass Spectrometry.” Bio Nano Frontier.
2013
- Sharma, Savita, Surekha M. Zingde, and Sadashiv M. Gokhale. 2013. “Identification of Human Erythrocyte Cytosolic Proteins Associated with Plasma Membrane during Thermal Stress.” Journal of Membrane Biology 246(8).
- Bhutada, Sumit, R. R. Katkam, Tarla Nandedkar, S. M. Metkari, U. K. Chaudhari, Sneha Varghese, S. D. Kholkute, and Geetanjali Sachdeva. 2013. “Uterine Secretome and Its Modulation in Rat (Rattus Norvegicus).” Reproduction 146(1).
- Kale, Avinash, Ramesh S. Hire, Ashok B. Hadapad, Stanislaus F. D’Souza, and Vinay Kumar. 2013. “Interaction between Mosquito-Larvicidal Lysinibacillus Sphaericus Binary Toxin Components: Analysis of Complex Formation.” Insect Biochemistry and Molecular Biology 43(11).
- Iyer, Sapna v., Prerana P. Dange, Hunain Alam, Sharada S. Sawant, Arvind D. Ingle, Anita M. Borges, Neelam v. Shirsat, Sorab N. Dalal, and Milind M. Vaidya. 2013. “Understanding the Role of Keratins 8 and 18 in Neoplastic Potential of Breast Cancer Derived Cell Lines.” PLoS ONE 8(1).
- Badgujar, Shamkant B., and Raghunath T. Mahajan. 2013. “ Peptide Mass Fingerprinting and N-Terminal Amino Acid Sequencing of Glycosylated Cysteine Protease of Euphorbia Nivulia Buch.-Ham. .” Journal of Amino Acids 2013
- Fulzele, A., S. A. Malgundkar, R. B. Govekar, A. Patil, S. V. Kane, P. Chaturvedi, A. K. D’Cruz, and S. M. Zingde. 2013. “Proteomic Profile of Keratins in Cancer of the Gingiva Buccal Complex: Consolidating Insights for Clinical Applications.” Journal of Proteomics 91:242–58.
2012
- Fulzele, Amit, Siddhi A. Malgundkar, Rukmini B. Govekar, Anil K. D’Cruz, Pankaj Chaturvedi, Asawari Patil, Shubhada v. Kane, and Surekha M. Zingde. 2012. “Keratins in Oral Cancer: Necessity of Mass Spectrometry for Validation of Antibody Based Identifications.” Journal of Proteomics 75(8).
- Kumar, Vinay, and Gagan D. Gupta. 2012. “Low-Resolution Structure of Drosophila Translin.” FEBS Open Bio 2
- Das, Sudipta, Rajagopal Sudarsan, Subramanian Sivakami, and Shobhona Sharma. 2012. “Erythrocytic Stage-Dependent Regulation of Oligomerization of Plasmodium Ribosomal Protein P2.” Journal of Biological Chemistry 287(49).
- Govekar, R., P. Kawle, R. Thomas, S. Advani, S. Pv, and S. Zingde. 2012. “Eryptotic Phenotype in Chronic Myeloid Leukemia: Contribution of Neutrophilic Cathepsin G. Anemia.” Anemia 659303(1 (2012)).
2011
- Khare, Satyajeet P., Ajitkumar Sharma, Kedar K. Deodhar, and Sanjay Gupta. 2011. “Overexpression of Histone Variant H2A.1 and Cellular Transformation Are Related in N-Nitrosodiethylamine-Induced Sequential Hepatocarcinogenesis.” Experimental Biology and Medicine 236(1).
- Chakraborty, Sandeep, Renu Minda, Lipika Salaye, Swapan K. Bhattacharjee, and Basuthkar J. Rao. 2011. “Active Site Detection by Spatial Conformity and Electrostatic Analysis-Unravelling a Proteolytic Function in Shrimp Alkaline Phosphatase.” PLoS ONE 6(12).
- Alam, Hunain, Samrat T. Kundu, Sorab N. Dalal, and Milind M. Vaidya. 2011. “Loss of Keratins 8 and 18 Leads to Alterations in Α6β4-Integrin-Mediated Signalling and Decreased Neoplastic Progression in an Oral-Tumour-Derived Cell Line.” Journal of Cell Science 124(12).
2009
- Parmar, Tanu, Sushama Gadkar-Sable, Lalita Savardekar, Rajendra Katkam, Shalmali Dharma, Pervin Meherji, Chander Parkash Puri, and Geetanjali Sachdeva. 2009. “Protein Profiling of Human Endometrial Tissues in the Midsecretory and Proliferative Phases of the Menstrual Cycle.” Fertility and Sterility 92(3).
- Pramod, Khare Satyajeet, Khade Bharat, and Gupta Sanjay. 2009. “Mass Spectrometry-Compatible Silver Staining of Histones Resolved on Acetic Acid-Urea-Triton PAGE.” Proteomics 9(9).
| Sr. No. | Name | Designation | Email ID/Phone/Extension |
|---|---|---|---|
| 1 | Mr. Shashadhar Dolas | Scientific Officer ‘E’ OIC-Mass Spectrometry Facility | Sdolas[at]actrec[dot]gov[dot]in 5475/5320 |
| 2 | Dr. Heramb Kulkarni | Scientific Officer ‘D’ | Heramb.kulkarni[at]actrec[dot]gov[dot]in 5475/5320/5394 |
| 3 | Mrs. Poonam Kawle | Scientific Assistant ‘F’ | mass.spec[at]actrec[dot]gov[dot]in 5475/5320 |
| 4 | Mr. Prince Ragul V. | Technician ’A’ | mass.spec[at]actrec[dot]gov[dot]in 5475/5320 |
Mass Spectrometry
KS-08 & KS-19, Khanolkar Shodhika,
Advanced Centre for Treatment, Research & Education in Cancer (ACTREC),
Tata Memorial Centre,
Sector-22, Kharghar,
Navi Mumbai – 410210,
Maharashtra, India.
Email : mass.spec[at]actrec[dot]gov[dot]in
Phone No: +91 (022) 27405000 / 68735000
Extension 5475/5320





